Introduction
automesh is an open-source Rust software program that uses a segmentation,
typically generated from a 3D image stack,
to create a finite element mesh,
composed either of hexahedral (volumetric)
or triangular (isosurface) elements.
automesh converts between
segmentation formats (.npy, .spn)
and
mesh formats (.exo, .inp, .mesh, .stl, .vtk).
automesh can defeature voxel domains,
apply Laplacian and Taubin smoothing,
and output mesh quality metrics.
automesh uses an internal octree for fast performance.
Segmentation
Segmentation is the process of categorizing pixels that compose a digital image into a class that represents some subject of interest. For example, in the image below, the image pixels are classified into classes of sky, trees, cat, grass, and cow.
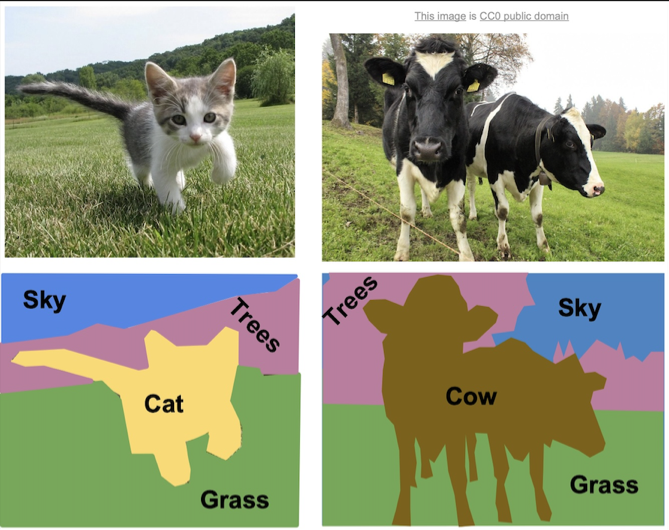
Figure: Example of semantic segmentation, from Li et al.1
- Semantic segmentation does not differentiate between objects of the same class.
- Instance segmentation does differentiate between objects of the same class.
These two concepts are shown below:
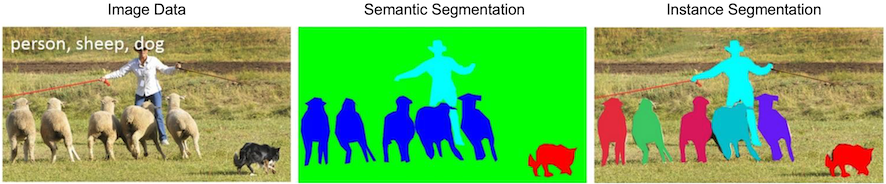
Figure: Distinction between semantic segmentation and instance segmentation, from Lin et al.2
Both segmentation types, semantic and instance, can be used with automesh. However, automesh operates on a 3D segmentation, not a 2D segmentation, as present in a digital image. To obtain a 3D segmentation, two or more images are stacked to compose a volume.
The structured volume of a stacked of pixel composes a volumetric unit called a voxel. A voxel, in the context of this work, will have the same dimensionality in the x and y dimension as the pixel in the image space, and will have the z dimensionality that is the stack interval distance between each image slice. All pixels are rectangular, and all voxels are cuboid.
The figure below illustrates the concept of stacked images:
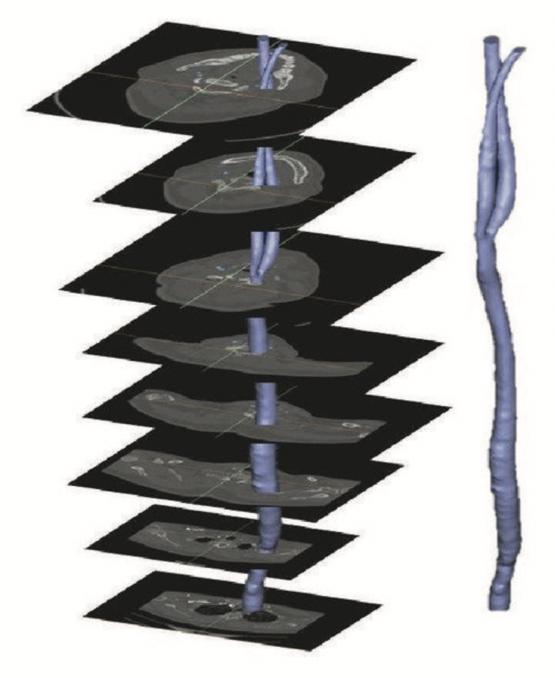
Figure: Example of stacking several images to create a 3D representation, from Bit et al.3
The digital image sources are frequently medical images, obtained by CT or MR, though automesh can be used for any subject that can be represented as a stacked segmentation. Anatomical regions are classified into categories. For example, in the image below, ten unique integers have been used to represent
bone, disc, vasculature, airway/sinus, membrane, cerebral spinal fluid, white matter, gray matter, muscle, and skin.
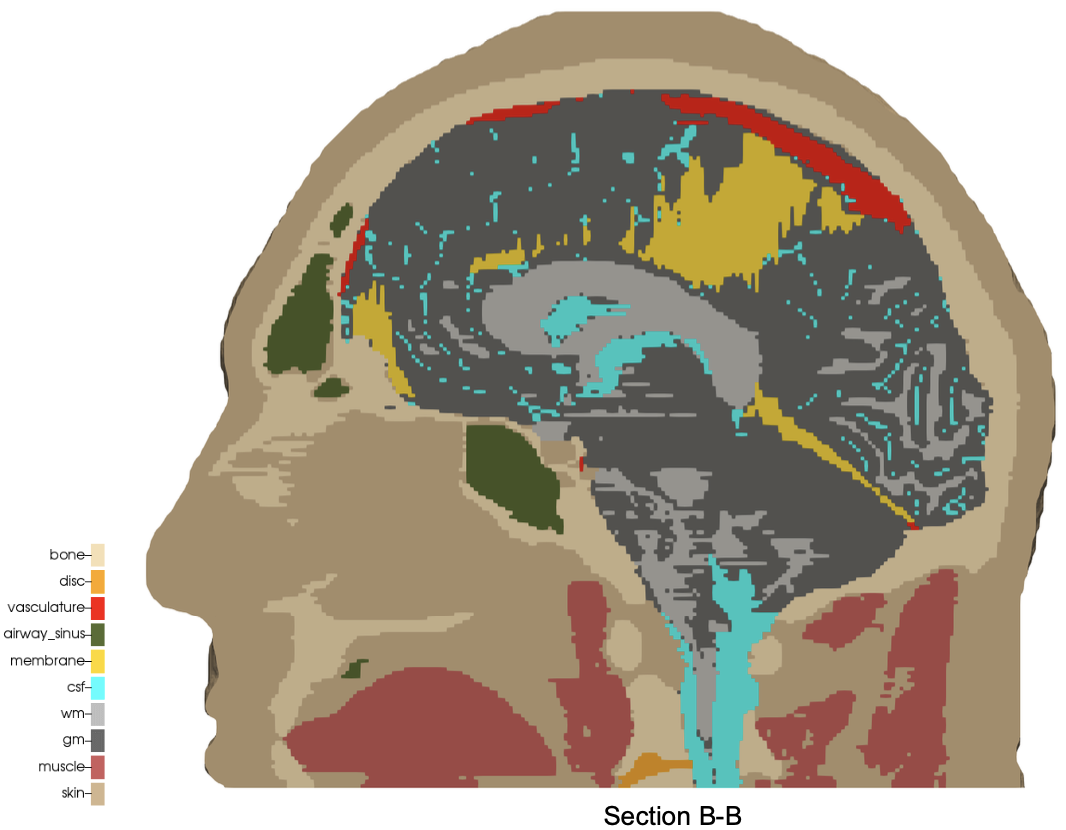
Figure: Example of a 3D voxel model, segmented into 10 categories, from Terpsma et al.4
Given a 3D segmentation, for any image slice that composes it, the pixels have been classified
into categories that are designated with unique, non-negative integers.
The range of integer values is limited to 256 = 2^8, since the uint8 data type is specified.
A practical example of a range could be [0, 1, 2, 3, 4]. The integers do not need to be sequential,
so a range of [4, 501, 2, 0, 42] is also valid, but not conventional.
Segmentations are frequently serialized (saved to disc) as either a NumPy (.npy) file
or a SPN (.spn) file.
A SPN file is a text (human-readable) file that contains a single a column of non-negative integer values. Each integer value defines a unique category of a segmentation.
Axis order (for example,
x, y, then z; or, z, y, x, etc.) is not implied by the SPN structure;
so additional data, typically provided through a configuration file, is
needed to uniquely interpret the pixel tile and voxel stack order
of the data in the SPN file.
For subjects that the human anatomy, we use the Patient Coordinate System (PCS), which directs the
x, y, and z axes to the left, posterior, and superior, as shown below:
| Patient Coordinate System: | Left, Posterior, Superior (x, y, z) |
|---|---|
 | 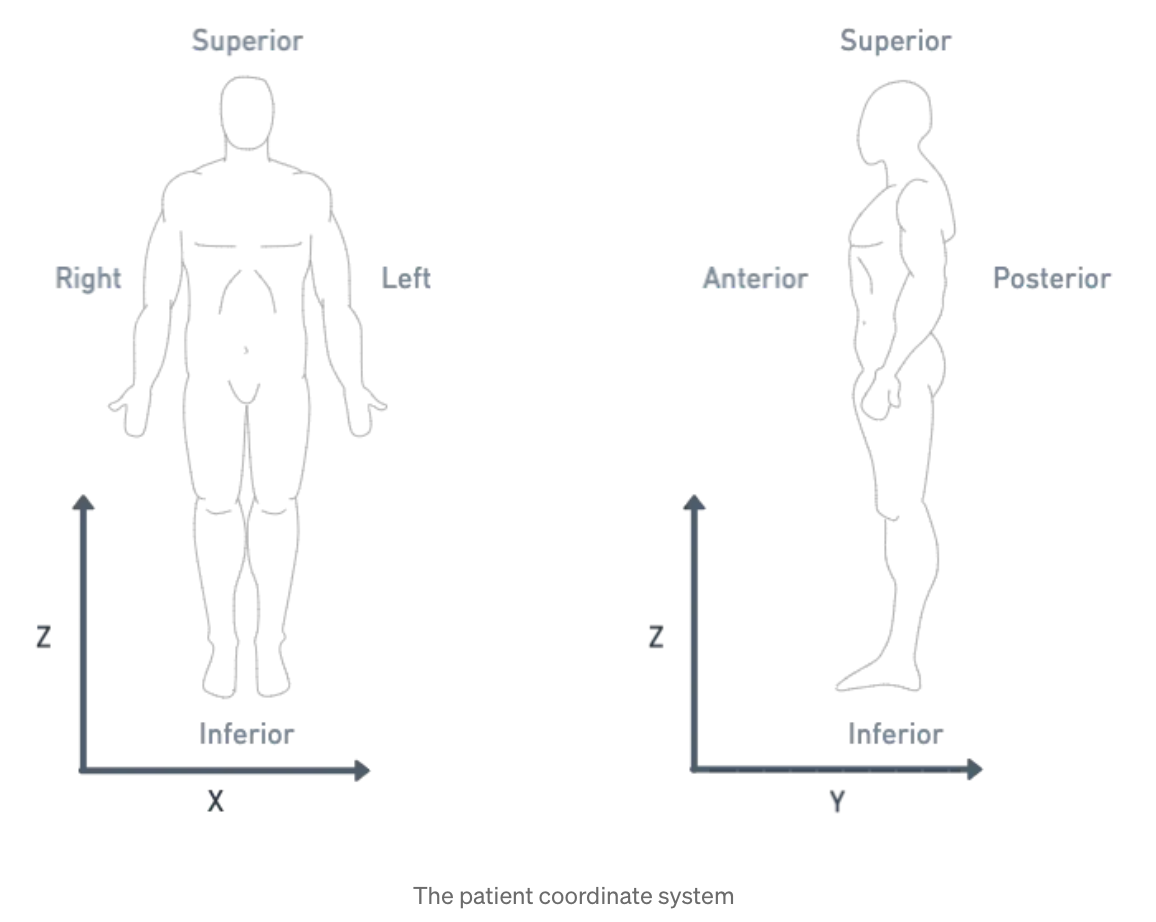 |
Figure: Illustration of the patient coordinate system, left figure from Terpsma et al.4 and right figure from Sharma.5
Finite Element Mesh
The main automesh file output type is .exo, which refers to the EXODUS II file format, or simply "Exodus" format.
Other mesh output types, such as .inp, .mesh, and .vtk, are also available.
EXODUS II is a model developed to store and retrieve data for finite element analyses. It is used for preprocesing (problem definition), postprocessing (results visualization), as well as code to code data transfer. An EXODUS II data file is a random access, machine independent binary file.6
EXODUS II depends on the Network Common Data Form (NetCDF) library.
NetCDF is a public domain database library that provides low-level data storage. The NetCDF library stores data in eXternal Data Representation (XDR) format, which provides machine independency.
EXODUS II library functions provide a map between finite element data objects and NetCDF dimensions, attributes, and variables.
EXODUS II data objects:
-
Initialization Data
- Number of nodes
- Number of elements
- optional informational text
- etc.
-
Model - static objects (i.e., objects that do not change over time)
- Nodal coordinates
- Element connectivity
- Node sets
- Side sets
-
optional Results
- Nodal results
- Element results
- Global results
Note:
automeshwill use Initialization Data and Model sections; it will not use the Results section.
We use the Exodus II convention for a hexahedral element local node numbering:
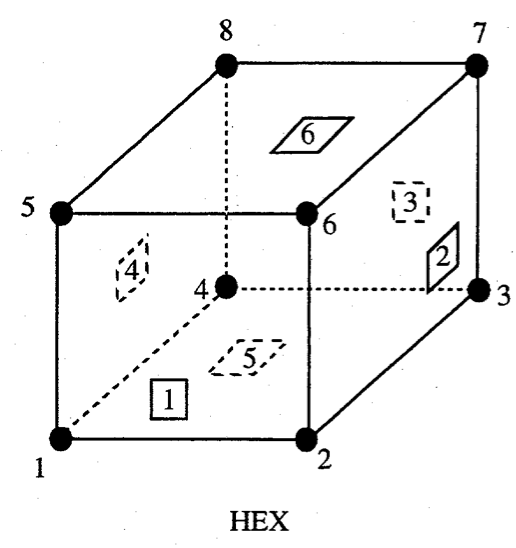
Figure: Exodus II hexahedral local finite element numbering scheme, from Schoof et al.6
References
-
Li FF, Johnson J, Yeung S. Lecture 11: Dection and Segmentation, CS 231n, Stanford Unveristy, 2017. link ↩
-
Lin TY, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, Zitnick CL. Microsoft coco: Common objects in context. In Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part V 13 2014 (pp. 740-755). Springer International Publishing. link ↩
-
Bit A, Ghagare D, Rizvanov AA, Chattopadhyay H. Assessment of influences of stenoses in right carotid artery on left carotid artery using wall stress marker. BioMed research international. 2017;2017(1):2935195. link ↩
-
Terpsma RJ, Hovey CB. Blunt impact brain injury using cellular injury criterion. Sandia National Lab. (SNL-NM), Albuquerque, NM (United States); 2020 Oct 1. link ↩ ↩2
-
Sharma S. DICOM Coordinate Systems — 3D DICOM for computer vision engineers, Medium, 2021-12-22. link ↩
-
Schoof LA, Yarberry VR. EXODUS II: a finite element data model. Sandia National Lab. (SNL-NM), Albuquerque, NM (United States); 1994 Sep 1. link ↩ ↩2