Cross-Correlation
Cross-correlation is a measure of similarity between two series, typically a time series. It is sometimes called the sliding dot product or sliding inner product.
The cross-correlation implements the following conceptual steps:
- Given two signals:
- is the reference signal with bounds .
- is the subject signal with bounds .
- Synchronization:
- Find the global minimum .
- Find the global maximum .
- Construct a global time interval .
- Choose a global time step, , to be the minimum of the reference time step and the subject time step.
- Correlation:
- Keep the reference signal stationary. Move the subject signal along the -axis until the last data point of the subject signal is multiplied by the first data point of the reference signal.
- Then, slide the subject signal to the right on the -axis by , calculating the inner product of the two signals for each in .
- Find the largest value of the foregoing inner products. Then for that step, move the subject curve to align with the reference curve. This will represent the highest correlation between the reference and the subject signal.
The figio implementation is based on the formulations contained in
Terpsma et al.1, Sections E.2 and E.3 (pages 110 to 121).
Example
Consider the sawtooth examples shown below, recreated from the Anomaly webpage, section Normalized Cross-Correlation with Time Shift.2
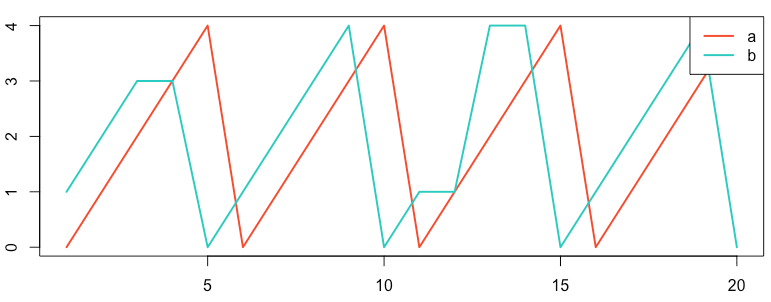
Figure: Reproduction of the sawtooth series on the Anomaly website.2
Create the input file anomaly_recipe.yml
::::::::::::::
anomaly_recipe.yml
::::::::::::::
signal_a:
type: xymodel
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_a.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
label: reference signal a
color: red
linewidth: 3
linestyle: "--"
marker: D
alpha: 0.9
signal_b:
type: xymodel
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_b.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
label: subject signal b
color: darkcyan
linewidth: 1
linestyle: "-"
marker: o
alpha: 0.8
signal_b_correlated:
type: xymodel
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_b.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
label: subject signal b
color: darkcyan
linewidth: 1
linestyle: "-"
marker: o
alpha: 0.8
signal_process:
process1:
correlation:
reference:
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_a.csv
skip_rows: 1
ycolumn: 1
verbose: true
serialize: false
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: out_signal_b_correlated.csv
figure_1:
type: xyview
model_keys: [ signal_a, signal_b ]
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: out_anomaly_pre_corr.svg
title: Anomaly site example, pre-correlation
xlabel: time (s)
ylabel: position (m)
xlim: [ -1, 22 ]
ylim: [ -1, 5 ]
size: [ 8.0, 6.0 ]
dpi: 100
display: true
details: false
serialize: true
figure_2:
type: xyview
model_keys: [ signal_a, signal_b_correlated ]
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: out_anomaly_post_corr.svg
title: Anomaly site example, post-correlation
xlabel: time (s)
ylabel: position (m)
xlim: [ -1, 22 ]
ylim: [ -1, 5 ]
size: [ 8.0, 6.0 ]
dpi: 100
display: true
details: false
serialize: true
which makes use of the two data series signal_a.csv and signal_b.csv.
::::::::::::::
signal_a.csv
::::::::::::::
time (s),signal_a (m)
1,0
2,1
3,2
4,3
5,4
6,0
7,1
8,2
9,3
10,4
11,0
12,1
13,2
14,3
15,4
16,0
17,1
18,2
19,3
20,4
::::::::::::::
signal_b.csv
::::::::::::::
time (s),signal_b (m)
1,1
2,2
3,3
4,3
5,0
6,1
7,2
8,3
9,4
10,0
11,1
12,1
13,4
14,4
15,0
16,1
17,2
18,3
19,4
20,0
Results
Run figio on the input file to produce the figures.
figio anomaly_recipe.yml
Processing file: anomaly_recipe.yml
====================================
Information
For (x, y) data and time series data:
type: xymodel items associate with type: xyview items.
For histogram data:
type: hmodel items associate with type: hview items.
====================================
This is xymodel.cross_correlation...
reference: [[ 1. 0.]
[ 2. 1.]
[ 3. 2.]
[ 4. 3.]
[ 5. 4.]
[ 6. 0.]
[ 7. 1.]
[ 8. 2.]
[ 9. 3.]
[10. 4.]
[11. 0.]
[12. 1.]
[13. 2.]
[14. 3.]
[15. 4.]
[16. 0.]
[17. 1.]
[18. 2.]
[19. 3.]
[20. 4.]]
subject: [[ 1. 1.]
[ 2. 2.]
[ 3. 3.]
[ 4. 3.]
[ 5. 0.]
[ 6. 1.]
[ 7. 2.]
[ 8. 3.]
[ 9. 4.]
[10. 0.]
[11. 1.]
[12. 1.]
[13. 4.]
[14. 4.]
[15. 0.]
[16. 1.]
[17. 2.]
[18. 3.]
[19. 4.]
[20. 0.]]
Synchronization...
Reference [t_min, t_max] by dt (s): [1.0, 20.0] by 1.0
Subject [t_min, t_max] by dt (s): [1.0, 20.0] by 1.0
Globalized [t_min, t_max] by dt (s): [1.0, 20.0] by 1.0
Globalized times: [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18.
19. 20.]
Length of globalized times: 20
Correlation...
Sliding dot product (cross-correlation): [ 0. 0. 4. 11. 20. 30. 20. 19. 27. 41. 61. 41. 30. 42.
61. 91. 61. 44. 55. 78. 117. 81. 51. 48. 58. 87. 61. 36.
32. 37. 56. 40. 25. 17. 17. 26. 20. 11. 4.]
Length of the sliding dot product: 39
Max sliding dot product (cross-correlation): 117.0
Sliding dot product of normalized signals (cross-correlation): [0. 0. 0.03375798 0.09283444 0.16878989 0.25318484
0.16878989 0.1603504 0.22786636 0.34601928 0.51480918 0.34601928
0.25318484 0.35445878 0.51480918 0.76799402 0.51480918 0.37133777
0.46417221 0.65828059 0.98742088 0.68359907 0.43041423 0.40509575
0.48949069 0.73423604 0.51480918 0.30382181 0.27006383 0.3122613
0.4726117 0.33757979 0.21098737 0.14347141 0.14347141 0.21942686
0.16878989 0.09283444 0.03375798]
Correlated time_shift (from full left)=20.0
Correlated index_shift (from full left)=20
Correlated time step (s): 1.0
Correlated t_min (s): 1.0
Correlated t_max (s): 21.0
Correlated times: [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18.
19. 20. 21.]
Correlated reference f(t): [0. 1. 2. 3. 4. 0. 1. 2. 3. 4. 0. 1. 2. 3. 4. 0. 1. 2. 3. 4. 0.]
Correlated subject f(t): [0. 1. 2. 3. 3. 0. 1. 2. 3. 4. 0. 1. 1. 4. 4. 0. 1. 2. 3. 4. 0.]
Correlated error f(t): [ 0. 0. 0. 0. -1. 0. 0. 0. 0. 0. 0. 0. -1. 1. 0. 0. 0. 0.
0. 0. 0.]
reference_self_correlation: 120.0
cross_correlation: 117.0
>> cross_correlation_relative_error=0.025
>> L2-norm error rate: 0.08247860988423225
Signal process "correlation" completed.
Finished XYViewBase constructor.
Finished XYViewBase constructor.
Creating view with guid = "figure_1"
Adding ['signal_a', 'signal_b'] model(s) to current view.
Figure dpi set to 100
Figure size set to [8.0, 6.0] inches.
Serialized view to: out_anomaly_pre_corr.svg
Creating view with guid = "figure_2"
Adding ['signal_a', 'signal_b_correlated'] model(s) to current view.
Figure dpi set to 100
Figure size set to [8.0, 6.0] inches.
Serialized view to: out_anomaly_post_corr.svg
====================================
End of figio execution.
Error metrics:
- cross-correlation relative error:
2.5 percent - L2-norm error rate:
8.3 percent
References
-
Terpsma RJ, Hovey CB. Blunt impact brain injury using cellular injury criterion. Sandia National Lab. (SNL-NM), Albuquerque, NM (United States); 2020 Oct 1. link ↩
-
Understanding Cross-Correlation, Auto-Correlation, Normalization and Time Shift, March 8, 2016. Available from: https://anomaly.io/understand-auto-cross-correlation-normalized-shift/ ↩ ↩2