Introduction
figio is a Python application that uses declarative YAML
input file recipes to produce high-quality matplotlib
and figures.
The following figure types are currently supported:
(x, y)data (or, equivalently, time series data), and- histogram data.
Installation
We support two types of installation: client or developer. The client installation is recommended for users who will use the module but are not interested in modifying the module's source code.
In contrast, the developer installation is recommended for users who wish to modify the module's source code.
Client Installation
Use of a virtual environment is recommended but not necessary.
python -m venv .venv
# Activate the venv with one of the following:
source .venv/bin/activate # for bash shell
source .venv/bin/activate.csh # for c shell
source .venv/bin/activate.fish # for fish shell
.\.venv\Scripts\activate # for powershell
Install figio from the Python Package Index (PyPI).
pip install figio
Getting Started
Tabular data is used as the data source for figio figures.
Let's get started with a tabular data for the inflation rate,
measured on the first day of each year.
The tabular data, inflation.csv, comes from the
Federal Reserve Bank of St. Louis.
::::::::::::::
inflation.csv
::::::::::::::
observation_date,FPCPITOTLZGUSA
1960,1.457975986277910
1961,1.070724147647240
1962,1.198773348201860
1963,1.239669421487530
1964,1.278911564625910
1965,1.585169263836620
1966,3.015075376884400
1967,2.772785622593090
1968,4.271796152885370
1969,5.462386200287450
1970,5.838255338482510
1971,4.292766688130510
1972,3.272278246552830
1973,6.177760063770380
1974,11.054804804804800
1975,9.143146864965351
1976,5.744812635490850
1977,6.501683994728400
1978,7.630963838856060
1979,11.254471129279500
1980,13.549201974968399
1981,10.334715340277100
1982,6.131427000274930
1983,3.212435233160650
1984,4.300535475234290
1985,3.545644152093650
1986,1.898047722342760
1987,3.664563217516900
1988,4.077741107444130
1989,4.827003030089440
1990,5.397956439903250
1991,4.234963964538490
1992,3.028819678149690
1993,2.951656966385590
1994,2.607441592154530
1995,2.805419688536620
1996,2.931204199934410
1997,2.337689937307350
1998,1.552279098743640
1999,2.188027196973580
2000,3.376857271499290
2001,2.826171118854070
2002,1.586031626506010
2003,2.270094973361150
2004,2.677236693091720
2005,3.392746845495500
2006,3.225944100704040
2007,2.852672481501380
2008,3.839100296651000
2009,-0.355546266299747
2010,1.640043442389900
2011,3.156841568622000
2012,2.069337265260670
2013,1.464832655627170
2014,1.622222977408170
2015,0.118627135552451
2016,1.261583205705360
2017,2.130110003659610
2018,2.442583296928170
2019,1.812210075260210
2020,1.233584396306290
2021,4.697858863637420
2022,8.002799820521210
2023,4.116338383744880
To plot this data, we create a yml input file called
inflation.yml.
::::::::::::::
inflation.yml
::::::::::::::
inflation-data:
type: xymodel
folder: ./ # the current directory
file: inflation.csv
skip_rows: 1
plot_kwargs:
label: inflation rate
linestyle: "-" # solid line
linewidth: 1.0
marker: "." # point marker
ycolumn: 1
inflation-figure:
type: xyview
folder: ./ # the current directory
file: inflation.svg # the output file
size: [ 8.0, 6.0 ]
title: Inflation, consumer prices for the United States (FPCPITOTLZGUSA)
xlabel: year
ylabel: inflation rate (%)
details: false
display: true
dpi: 100
latex: false
serialize: true
Run figio on the input file to produce the figure:
figio inflation.yml
Processing file: inflation.yml
====================================
Information
For (x, y) data and time series data:
type: xymodel items associate with type: xyview items.
For histogram data:
type: hmodel items associate with type: hview items.
====================================
Finished XYViewBase constructor.
Creating view with guid = "inflation-figure"
Adding all models to current view.
Figure dpi set to 100
Figure size set to [8.0, 6.0] inches.
Serialized view to: inflation.svg
====================================
End of figio execution.
The following figure appears:
Congratulations! You just made your first figio figure.
Keys and Values
Below are dictionary key: value pairs, followed by a description, for each
of the figio dictionary constitutents.
Main figio Dictionary
The figio dictionary is the main dictionary.
- It is stored in an ordinary text file in
ymlformat. - It is composed of one or more
modeldictionaries and one or moreviewdictionaries.
model_name: | dict | The key model_name is a string that must be a globally unique identifier (guid) in the yml file. Contains the model dictionary. Non-singleton; supports 1..m models. |
view_name: | dict | The key view_name is a string that must be a globally unique identifier (guid) in the yml file. The key Contains the view dictionary. Non-singleton, supports 1..n views.Note: In general, this view_name key can be any unique string. However, when the yml input file is to be used with the unit tests, this view_name key string must be exactly set to figure for the unit tests to work properly. |
There are three common variations to the figio dictionary:
- 1:1
- One model
- One view
- This is the most basic option.
- 1:n
- Several models
- One view
- This is an advanced option, wherein multiple models are plotted to the same figure. Considerations such as y-axis limits or the possibility of dual y-axes can become important.
- m:n
- Several models
- Several views
- This is the most advanced combination of the three, where all the models are created, and views explicity specify which models get plotted to a particular figure.
Signal processing may be performed on one or more models, using the signal_process dictionary, to create a new model, which can also be used by the view. A conceptual flow diagram of multiple models, one with signal processing, and one view is shown below.
┌───────────────┐ ┌───────────────┐
│ Model │─────────────────────────┐ │ │
└───────────────┘ │ │ │
│ │ │
┌───────────────┐ │ │ │
│ Model │─────────────────────────┤ │ │
└───────────────┘ │ │ │
│ │ │ View │
├─────────────────────┬───▶│ │
│ │ │ │ │
┌───────────────┐ │ │ │ │
│ Model │──┬──────────────────────┘ │ │ │
└───────────────┘ │ ┌───────────────┐ │ │ │
│ │ Signal │ ┌───────────────┐ │ │ │
└──▶│ Process │───▶│ Model │───┘ │ │
└───────────────┘ └───────────────┘ └───────────────┘
Model Dictionary
The model dictionary contains items that describe how each (x,y) data set is
constructed and shown on the view.
type: | xymodel hmodel | For (x, y) data and time series data, type: xymodel items associate with type: xyview items. For histogram data, type: hmodel items associate with type: hview items. |
folder: | string | Value of the absolute file path to the file. Supports ~ for user home constructs ~/my_project/input_files as equilvalent to, for example, /Users/chovey/my_project/input_files. For the current working directory, use ./. |
file: | string | Value of the comma separated value input file in csv (comma separated value) format. The first column is the x values, the second column(s) is(are) the y value(s). The csv file can use any number of header rows. Do not attempt to plot header rows; skip header rows with the skip_rows key. |
skip_rows: | integer | optional The number of header rows to skip at the beginning of the csv file. Default value is 0. |
skip_rows_footer: | integer | optional The number of footer rows to skip at the end of the csv file. Default value is 0. |
xcolumn: | integer | optional The zero-based index of the data column to plotted on the x-axis. Default is 0, which is the first column of the csv file. |
ycolumn: | integer | optional The zero-based index of the data column to be plotted on the y-axis. Default is 1, which is the second column of the csv file. |
xscale: | float | optional Scales all values of the x data xscale factor. Default value is 1.0 (no scaling). xscale is applied to the data prior to xoffset. |
xoffset: | float | optional Shifts all values of the x data to the left or the right by the xoffset value. Default value is 0.0. xoffset is applied to the data after xscale. |
yscale: | float | optional Scales all values of the y data yscale factor. Default value is 1.0 (no scaling). yscale is applied to the data prior to yoffset. |
yoffset: | float | optional Shifts all values of the y data up or down by the yoffset value. Default value is 0.0. yoffset is applied to the data after yscale. |
plot_kwargs: | dict | Singleton that contains the plot keywords dictionary. |
signal_process: | dict | Singleton that contains the signal processing keywords dictionary. |
Plot Keywords Dictionary
Dictionary that overrides the matplotlib.pyplot.plot() kwargs default values. Default values used by figio follow:
linewidth: | float | optional Default value is 2.0. See matplotlib lines Line2D for more detail. |
linestyle: | string | optional Default value is "-", which is a solid line. See matplotlib linestyles for more detail. |
Some frequently used optional values follow. If the keys are omitted, then the matplotlib defaults are used. | ||
marker: | string | optional The string to designate a marker at the data point. See matplotlib marker documentation. |
label: | string | optional The string appearing in the legend correponding to the data. |
color: | string | optional The matplotlib color used to plot the data. See also matplotlib color defaults and predefined color names. |
alpha: | float | optional Real number in the range from 0.0 to 1.0. Numbers toward 0.0 are more transparent and numbers toward 1.0 are more opaque. |
Signal Processing Keywords Dictionary
Warning: The Signal Processing dictionary and links below are a work in progress.
This dictionary is currently under active development. For additional documentation, see
- Butterworth filter
- Differentiation
- Integration
- Cross correlation (to come, not yet implemented)
- tpav (three points angular velocity) algorithm
Below is a summary of the key: value pairs available within the signal_process dictionary.
signal_process:
process_guid_1:
butterworth:
cutoff: 5
order: 4
type: low
process_guid_2:
gradient:
order: 1
process_guid_3:
integration:
order: 3
initial_conditions: [-10, 100, 1000]
process_guid_4:
crosscorrelation:
model_keys: [model_guid_0, model_guid_1]
mode: full # (valid | same)
process_guid_5:
tpav: { (tpav is three points angular velocity)
model_keys: [model_guid_0, model_guid_1, model_guid_2]
All processes support serialization, via
signal_process:
process_guid:
process_key_string:
serialize: 1
folder: ~/sibl/cli/io/example
file: processed_output_file.csv
View Dictionary
The view dictionary contains items that describe how the main figure is constructed.
type: | xyview hview | For (x, y) data and time series data, type: xymodel items associate with type: xyview items. For histogram data, type: hmodel items associate with type: hview items. |
model_keys: | string array | optional[model_guid_0, model_guid_1, model_guid_2] (for example), an array of 1..m strings that identifies the model guid to be plotted in this particular view. If model_keys is not specified in a particular view, then all models will be plotted to the view, which is the default behavior. |
folder: | string | Value of the absolute file path to the file. Supports ~ for user home constructs ~/my_project/output_files as equilvalent to, for example, /Users/chovey/my_project/output_files. |
file: | string | Value of the figure output file (e.g., my_output_file.png) in .xxx format, where xxx is an image file format, typically pdf, png, or svg. |
size: | float array | optional Array of floats containing the [width, height] of the output figure in units of inches. Default is [11.0, 8.5], U.S. paper, landscape. Example |
dpi: | integer | optional Dots per inch used for the output figure. Default is 300. Example |
xlim: | float array | optional Array of floats containing the x-axis bounds [x_min, x_max]. Default is MATLABLIB's automatic selection. |
ylim: | float array | optional Array of floats containing the y-axis bounds [y_min, y_max]. Default is matplotlib's automatic selection. |
title: | string | optional Figure label, top and centered. Default is default title. |
xlabel: | string | optional The label for the x-axis. Default is default x axis label. |
ylabel: | string | optional The label for the left-hand y-axis. Default is default y axis label. |
frame: | Boolean | optional Visibility of the rectangular frame draw around an axis. 0 The frame is hidden.1 (default) The frame is shown. |
tick_kwargs: | dict | optional Singleton that controls ticks, tick labels, and gridlines per matplotlib's tick_params dictionary.Example: tick_params: {colors: darkgray, labelsize: 8, width: 0.1} |
xticks: | float array | optional Contains an array of ascending real numbers, indicating tick placement. Example: [0.0, 0.5, 1.0, 1.5, 2.0]. Default is matplotlib's choice for tick marks. |
yticks: | float array | optional Same as documentation for xticks. |
xaxis_log: | Boolean | optional0 (default) plots x-axis in a linear scale.1 plots x-axis in a log scale. |
yaxis_log: | Boolean | optional0 (default) plots y-axis in a linear scale.1 plots y-axis in a log scale. |
yaxis_rhs: | dict | optional Singleton that contains the yaxis_rhs dictionary. |
background_image: | dict | optional Singleton that contains the background_image dictionary. |
display: | Boolean | optional0 to suppress showing figure in GUI, useful when serializing multiple figures during a parameter search loop.1 (default) to show figure interactively, and to pause script execution. |
latex: | string | optional0 (default) uses matplotlib default fonts, results in fast generation of figures.1 uses LaTeX fonts, can be slow to generate, but produces production-quality results. |
details: | Boolean | optional0 do not show plot details.1 (default) shows plot details of figure file name, date (yyyy-mm-dd format), and time (hh:mm:ss format) the figure was generated, and username. |
serialize: | string | optional0 (default) does not save figure to the file system.1 saves figure to the file system. Tested on local drives, but not on network drives. |
yaxis_rhs Dictionary
scale: | string | optional The factor that multiplies the left-hand y-axis to produce the right-hand y-axis. For example, if the left-hand y-axis is in meters, and the right-hand y-axis is in centimeters, the value of scale should be set to 100. Default value is 1. |
label: | string | optional The right-hand-side y-axis label. Default is an empty string ( None). |
yticks: | float array | optional Same as documentation for xticks. |
background_image Dictionary
folder: | string | Value relative to the current working directory of the path and folder that contains the background image file. For the current working directory, use ./. |
file: | string | Value of the background image file name, typically in png format. |
left: | float | optional Left-side extent of image in plot x coordinates. Must be less than the right-side extent. Default is 0.0. |
right: | float | optional Right-side extent of image in plot x coordinates. Must be greater than the left-side extent. Default is 1.0. |
bottom: | float | optional Bottom-side extent of image in plot y coordinates. Must be less than the top-side extent. Default is 0.0. |
top: | float | optional Top-side extent of image in plot y coordinates. Must be greater than the bottom-side extent. Default is 1.0. |
alpha: | float | optional Real number in the range from 0 to 1. Numbers toward 0 are more transparent and numbers toward 1 are more opaque. Default is 1.0 (fully opaque, no transparency). |
Multiple Series
We show an example of multiple series using default and non-default values for the line specifications (color, transparency, and type).
The file data.csv
contains columns for time (s),sin(t),cos(2t).
::::::::::::::
data.csv
::::::::::::::
time (s),sin(t),cos(2t)
0,0,1
0.1,0.099833417,0.980066578
0.2,0.198669331,0.921060994
0.3,0.295520207,0.825335615
0.4,0.389418342,0.696706709
0.5,0.479425539,0.540302306
0.6,0.564642473,0.362357754
0.7,0.644217687,0.169967143
0.8,0.717356091,-0.029199522
0.9,0.78332691,-0.227202095
1,0.841470985,-0.416146837
1.1,0.89120736,-0.588501117
1.2,0.932039086,-0.737393716
1.3,0.963558185,-0.856888753
1.4,0.98544973,-0.942222341
1.5,0.997494987,-0.989992497
1.6,0.999573603,-0.998294776
1.7,0.99166481,-0.966798193
1.8,0.973847631,-0.896758416
1.9,0.946300088,-0.790967712
2,0.909297427,-0.653643621
2.1,0.863209367,-0.490260821
2.2,0.808496404,-0.30733287
2.3,0.745705212,-0.112152527
2.4,0.675463181,0.087498983
2.5,0.598472144,0.283662185
2.6,0.515501372,0.468516671
2.7,0.42737988,0.634692876
2.8,0.33498815,0.775565879
2.9,0.239249329,0.885519517
3,0.141120008,0.960170287
3.1,0.041580662,0.996542097
3.2,-0.058374143,0.993184919
3.3,-0.157745694,0.950232592
3.4,-0.255541102,0.86939749
3.5,-0.350783228,0.753902254
3.6,-0.442520443,0.608351315
3.7,-0.529836141,0.438547328
3.8,-0.611857891,0.251259843
3.9,-0.687766159,0.053955421
4,-0.756802495,-0.145500034
4.1,-0.818277111,-0.339154861
4.2,-0.871575772,-0.519288654
4.3,-0.916165937,-0.678720047
4.4,-0.951602074,-0.811093014
4.5,-0.977530118,-0.911130262
4.6,-0.993691004,-0.974843621
4.7,-0.999923258,-0.999693042
4.8,-0.996164609,-0.984687856
4.9,-0.982452613,-0.930426272
5,-0.958924275,-0.839071529
5.1,-0.925814682,-0.714265652
5.2,-0.883454656,-0.560984257
5.3,-0.832267442,-0.385338191
5.4,-0.772764488,-0.194329906
5.5,-0.705540326,0.004425698
5.6,-0.631266638,0.203004864
5.7,-0.550685543,0.393490866
5.8,-0.464602179,0.56828963
5.9,-0.373876665,0.720432479
6,-0.279415498,0.843853959
6.1,-0.182162504,0.933633644
6.2,-0.083089403,0.986192302
6.3,0.0168139,0.999434586
To plot this data, we create a yml input file called
recipe.yml.
::::::::::::::
recipe.yml
::::::::::::::
sine-data:
type: xymodel
folder: ./ # the current directory
file: data.csv
skip_rows: 1
plot_kwargs:
label: sin(t)
color: darkorange
linestyle: "-"
linewidth: 4
alpha: 0.5
cosine-data:
type: xymodel
folder: ./ # the current directory
file: data.csv
skip_rows: 1
ycolumn: 2
plot_kwargs:
label: cos(2t)
linestyle: "-."
linewidth: 2
figure:
type: xyview
folder: ./ # the current directory
file: recipe.svg
size: [ 8.0, 6.0 ]
title: Example using figio
xlabel: time (s)
ylabel: functions of time f(t)
yaxis_rhs:
scale: 10
label: right-hand-side label with 10 f(t) scale
yticks: [ -10, -5, 0, 5, 10 ]
details: false
display: true
dpi: 100
latex: false
serialize: true
Run figio on the input file to produce the figure:
figio recipe.yml
Processing file: recipe.yml
====================================
Information
For (x, y) data and time series data:
type: xymodel items associate with type: xyview items.
For histogram data:
type: hmodel items associate with type: hview items.
====================================
Finished XYViewBase constructor.
Creating view with guid = "figure"
Adding all models to current view.
Figure dpi set to 100
Figure size set to [8.0, 6.0] inches.
Serialized view to: recipe.svg
====================================
End of figio execution.
The following figure appears:
Congratulations! You just made a figio figure with two data sources.
Cross-Correlation
Cross-correlation is a measure of similarity between two series, typically a time series. It is sometimes called the sliding dot product or sliding inner product.
The cross-correlation implements the following conceptual steps:
- Given two signals:
- is the reference signal with bounds .
- is the subject signal with bounds .
- Synchronization:
- Find the global minimum .
- Find the global maximum .
- Construct a global time interval .
- Choose a global time step, , to be the minimum of the reference time step and the subject time step.
- Correlation:
- Keep the reference signal stationary. Move the subject signal along the -axis until the last data point of the subject signal is multiplied by the first data point of the reference signal.
- Then, slide the subject signal to the right on the -axis by , calculating the inner product of the two signals for each in .
- Find the largest value of the foregoing inner products. Then for that step, move the subject curve to align with the reference curve. This will represent the highest correlation between the reference and the subject signal.
The figio implementation is based on the formulations contained in
Terpsma et al.1, Sections E.2 and E.3 (pages 110 to 121).
Example
Consider the sawtooth examples shown below, recreated from the Anomaly webpage, section Normalized Cross-Correlation with Time Shift.2
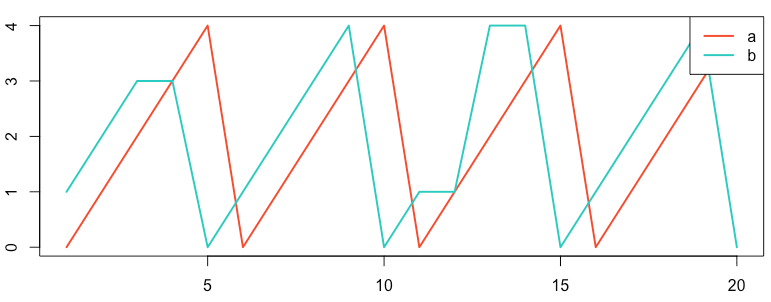
Figure: Reproduction of the sawtooth series on the Anomaly website.2
Create the input file anomaly_recipe.yml
::::::::::::::
anomaly_recipe.yml
::::::::::::::
signal_a:
type: xymodel
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_a.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
label: reference signal a
color: red
linewidth: 3
linestyle: "--"
marker: D
alpha: 0.9
signal_b:
type: xymodel
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_b.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
label: subject signal b
color: darkcyan
linewidth: 1
linestyle: "-"
marker: o
alpha: 0.8
signal_b_correlated:
type: xymodel
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_b.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
label: subject signal b
color: darkcyan
linewidth: 1
linestyle: "-"
marker: o
alpha: 0.8
signal_process:
process1:
correlation:
reference:
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: signal_a.csv
skip_rows: 1
ycolumn: 1
verbose: true
serialize: false
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: out_signal_b_correlated.csv
figure_1:
type: xyview
model_keys: [ signal_a, signal_b ]
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: out_anomaly_pre_corr.svg
title: Anomaly site example, pre-correlation
xlabel: time (s)
ylabel: position (m)
xlim: [ -1, 22 ]
ylim: [ -1, 5 ]
size: [ 8.0, 6.0 ]
dpi: 100
display: true
details: false
serialize: true
figure_2:
type: xyview
model_keys: [ signal_a, signal_b_correlated ]
# folder: ~/autotwin/figio/book/cross_correlation
folder: ./ # the current directory
file: out_anomaly_post_corr.svg
title: Anomaly site example, post-correlation
xlabel: time (s)
ylabel: position (m)
xlim: [ -1, 22 ]
ylim: [ -1, 5 ]
size: [ 8.0, 6.0 ]
dpi: 100
display: true
details: false
serialize: true
which makes use of the two data series signal_a.csv and signal_b.csv.
::::::::::::::
signal_a.csv
::::::::::::::
time (s),signal_a (m)
1,0
2,1
3,2
4,3
5,4
6,0
7,1
8,2
9,3
10,4
11,0
12,1
13,2
14,3
15,4
16,0
17,1
18,2
19,3
20,4
::::::::::::::
signal_b.csv
::::::::::::::
time (s),signal_b (m)
1,1
2,2
3,3
4,3
5,0
6,1
7,2
8,3
9,4
10,0
11,1
12,1
13,4
14,4
15,0
16,1
17,2
18,3
19,4
20,0
Results
Run figio on the input file to produce the figures.
figio anomaly_recipe.yml
Processing file: anomaly_recipe.yml
====================================
Information
For (x, y) data and time series data:
type: xymodel items associate with type: xyview items.
For histogram data:
type: hmodel items associate with type: hview items.
====================================
This is xymodel.cross_correlation...
reference: [[ 1. 0.]
[ 2. 1.]
[ 3. 2.]
[ 4. 3.]
[ 5. 4.]
[ 6. 0.]
[ 7. 1.]
[ 8. 2.]
[ 9. 3.]
[10. 4.]
[11. 0.]
[12. 1.]
[13. 2.]
[14. 3.]
[15. 4.]
[16. 0.]
[17. 1.]
[18. 2.]
[19. 3.]
[20. 4.]]
subject: [[ 1. 1.]
[ 2. 2.]
[ 3. 3.]
[ 4. 3.]
[ 5. 0.]
[ 6. 1.]
[ 7. 2.]
[ 8. 3.]
[ 9. 4.]
[10. 0.]
[11. 1.]
[12. 1.]
[13. 4.]
[14. 4.]
[15. 0.]
[16. 1.]
[17. 2.]
[18. 3.]
[19. 4.]
[20. 0.]]
Synchronization...
Reference [t_min, t_max] by dt (s): [1.0, 20.0] by 1.0
Subject [t_min, t_max] by dt (s): [1.0, 20.0] by 1.0
Globalized [t_min, t_max] by dt (s): [1.0, 20.0] by 1.0
Globalized times: [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18.
19. 20.]
Length of globalized times: 20
Correlation...
Sliding dot product (cross-correlation): [ 0. 0. 4. 11. 20. 30. 20. 19. 27. 41. 61. 41. 30. 42.
61. 91. 61. 44. 55. 78. 117. 81. 51. 48. 58. 87. 61. 36.
32. 37. 56. 40. 25. 17. 17. 26. 20. 11. 4.]
Length of the sliding dot product: 39
Max sliding dot product (cross-correlation): 117.0
Sliding dot product of normalized signals (cross-correlation): [0. 0. 0.03375798 0.09283444 0.16878989 0.25318484
0.16878989 0.1603504 0.22786636 0.34601928 0.51480918 0.34601928
0.25318484 0.35445878 0.51480918 0.76799402 0.51480918 0.37133777
0.46417221 0.65828059 0.98742088 0.68359907 0.43041423 0.40509575
0.48949069 0.73423604 0.51480918 0.30382181 0.27006383 0.3122613
0.4726117 0.33757979 0.21098737 0.14347141 0.14347141 0.21942686
0.16878989 0.09283444 0.03375798]
Correlated time_shift (from full left)=20.0
Correlated index_shift (from full left)=20
Correlated time step (s): 1.0
Correlated t_min (s): 1.0
Correlated t_max (s): 21.0
Correlated times: [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18.
19. 20. 21.]
Correlated reference f(t): [0. 1. 2. 3. 4. 0. 1. 2. 3. 4. 0. 1. 2. 3. 4. 0. 1. 2. 3. 4. 0.]
Correlated subject f(t): [0. 1. 2. 3. 3. 0. 1. 2. 3. 4. 0. 1. 1. 4. 4. 0. 1. 2. 3. 4. 0.]
Correlated error f(t): [ 0. 0. 0. 0. -1. 0. 0. 0. 0. 0. 0. 0. -1. 1. 0. 0. 0. 0.
0. 0. 0.]
reference_self_correlation: 120.0
cross_correlation: 117.0
>> cross_correlation_relative_error=0.025
>> L2-norm error rate: 0.08247860988423225
Signal process "correlation" completed.
Finished XYViewBase constructor.
Finished XYViewBase constructor.
Creating view with guid = "figure_1"
Adding ['signal_a', 'signal_b'] model(s) to current view.
Figure dpi set to 100
Figure size set to [8.0, 6.0] inches.
Serialized view to: out_anomaly_pre_corr.svg
Creating view with guid = "figure_2"
Adding ['signal_a', 'signal_b_correlated'] model(s) to current view.
Figure dpi set to 100
Figure size set to [8.0, 6.0] inches.
Serialized view to: out_anomaly_post_corr.svg
====================================
End of figio execution.
Error metrics:
- cross-correlation relative error:
2.5 percent - L2-norm error rate:
8.3 percent
References
-
Terpsma RJ, Hovey CB. Blunt impact brain injury using cellular injury criterion. Sandia National Lab. (SNL-NM), Albuquerque, NM (United States); 2020 Oct 1. link ↩
-
Understanding Cross-Correlation, Auto-Correlation, Normalization and Time Shift, March 8, 2016. Available from: https://anomaly.io/understand-auto-cross-correlation-normalized-shift/ ↩ ↩2
Histograms
Given a particular column from a csv data source, figio can create a frequency count histogram.
We illustrate this functionality on a large data set called sr2s10.csv (5.4 MB) and sr2s50.csv (11 MB).
To reproduce this example, download the two data sets and place them in a local ~/temp folder.
For concreteness in the following discussion, the first 10 rows of sr2s10.csv appear below:
maximum edge ratio, minimum scaled jacobian, maximum skew, volume
1.000000e0, 6.663165e-1, 2.872659e-1, 4.368155e-2
1.193120e0, 6.478237e-1, 4.187394e-1, 8.360088e-2
1.247881e0, 6.474025e-1, 4.065007e-1, 9.447332e-2
1.214427e0, 6.549605e-1, 3.901043e-1, 8.909662e-2
1.179123e0, 6.668604e-1, 3.859307e-1, 8.359763e-2
1.171296e0, 6.702009e-1, 3.868204e-1, 8.222564e-2
1.175480e0, 6.695364e-1, 3.874507e-1, 8.270613e-2
1.178150e0, 6.694542e-1, 3.873941e-1, 8.308343e-2
To plot this data, we create a yml input file called histogram.yml.
::::::::::::::
histogram.yml
::::::::::::::
s10_aspect:
type: hmodel
folder: ~/temp
file: sr2s10.csv
skip_rows: 1
ycolumn: 0
plot_kwargs:
alpha: 0.7
color: blue
label: sr2s10
linewidth: 2.0
s10_msj:
type: hmodel
folder: ~/temp
file: sr2s10.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
alpha: 0.7
color: blue
label: sr2s10
linewidth: 2.0
s10_skew:
type: hmodel
folder: ~/temp
file: sr2s10.csv
skip_rows: 1
ycolumn: 2
plot_kwargs:
alpha: 0.7
color: blue
label: sr2s10
linewidth: 2.0
s10_vol:
type: hmodel
folder: ~/temp
file: sr2s10.csv
skip_rows: 1
ycolumn: 3
plot_kwargs:
alpha: 0.7
color: blue
label: sr2s10
linewidth: 2.0
s50_aspect:
type: hmodel
folder: ~/temp
file: sr2s50.csv
skip_rows: 1
ycolumn: 0
plot_kwargs:
alpha: 0.7
color: orange
label: sr2s50
linewidth: 2.0
s50_msj:
type: hmodel
folder: ~/temp
file: sr2s50.csv
skip_rows: 1
ycolumn: 1
plot_kwargs:
alpha: 0.7
color: orange
label: sr2s50
linewidth: 2.0
s50_skew:
type: hmodel
folder: ~/temp
file: sr2s50.csv
skip_rows: 1
ycolumn: 2
plot_kwargs:
alpha: 0.7
color: orange
label: sr2s50
linewidth: 2.0
s50_vol:
type: hmodel
folder: ~/temp
file: sr2s50.csv
skip_rows: 1
ycolumn: 3
plot_kwargs:
alpha: 0.7
color: orange
label: sr2s50
linewidth: 2.0
figure_aspect:
type: hview
models: [s10_aspect, s50_aspect]
folder: ./ # the current directory
file: hist_sr2sx_aspect.svg
size: [8.0, 6.0]
title: Maximum Aspect Ratio
xlabel: Maximum Aspect Ratio
ylabel: Frequency
details: true
display: true
dpi: 100
latex: false
save: true
figure_msj:
type: hview
models: [s10_msj, s50_msj]
folder: ./ # the current directory
file: hist_sr2sx_msj.svg
size: [8.0, 6.0]
title: Minimum Scaled Jacobian
xlabel: Minimum Scaled Jacobian
ylabel: Frequency
details: true
display: true
dpi: 100
latex: false
save: true
figure_skew:
type: hview
models: [s10_skew, s50_skew]
folder: ./ # the current directory
file: hist_sr2sx_skew.svg
size: [8.0, 6.0]
title: Maximum Skew
xlabel: Maximum Skew
ylabel: Frequency
details: true
display: true
dpi: 100
latex: false
save: true
figure_vol:
type: hview
models: [s10_vol, s50_vol]
folder: ./ # the current directory
file: hist_sr2sx_vol.svg
size: [8.0, 6.0]
title: Element Volume
xlabel: Element Volume
ylabel: Frequency
details: true
display: true
dpi: 100
latex: false
save: true
Run figio on the input file to produce the figures:
figio histogram.yml
Processing file: histogram.yml
====================================
Information
For (x, y) data and time series data:
type: xymodel items associate with type: xyview items.
For histogram data:
type: hmodel items associate with type: hview items.
====================================
Histogram default plot_kwargs: {'alpha': 1.0, 'bins': 20, 'color': 'black', 'histtype': 'step', 'label': '', 'linewidth': 1.0, 'log': True}
Histogram updated plot_kwargs: {'alpha': 0.7, 'bins': 20, 'color': 'blue', 'histtype': 'step', 'label': 'sr2s10', 'linewidth': 2.0, 'log': True}
Valid: Validated data: {'alpha': 0.7, 'bins': 20, 'color': 'blue', 'histtype': 'step', 'label': 'sr2s10', 'linewidth': 2.0, 'log': True}
The following figures appear:
Development
Client Configuration
pip install figio
Developer Configuration
Since figio is a package within the autotwin framework, we suggest cloning the
figio repo to the autotwin folder:
cd ~/autotwin
git clone git@github.com:autotwin/figio.git
cd ~/autotwin/figio
From the ~/autotwin/figio folder, create the virtual environment.
A virtual environment is a self-contained directory that contains a specific Python
installation, along with additional packages. It allows users to create isolated
environments for different projects. This ensures that dependencies and libraries
do not interfere with each other.
Create a virtual environment with either pip or uv. pip is already included
with Python. uv must be installed.
uv is 10-100x faster than pip.
# pip method
python -m venv .venv
# uv method
uv venv
# both methods
source .venv/bin/activate # bash
source .venv/bin/activate.fish # fish shell
Install the code in editable form,
# pip method
pip install -e .[dev]
# uv method
uv pip install -e .[dev]
Manual Distribution
python -m build . --sdist # source distribution
python -m build . --wheel
twine check dist/*
Distribution
The distribution steps will tag the code state as a release version, with a semantic version number, build the code as a wheel file, and publish the wheel file as a release to GitHub.
Tag
View existing tags, if any:
git tag
Create a tag. Tags can be lightweight or annotated. Annotated tags are recommended since they store tagger name, email, date, and message information. Create an annotated tag:
# example of an annotated tag
git tag -a v1.0.0 -m "Release version 1.0.0"
Push the tag to the repo
# example continued
git push origin v1.0.0
Verify the tag appears on the repo
Build
Ensure that setuptools and build are installed:
pip install setuptools build
Navigate to the project directory, where the pyproject.toml file is located,
and create a wheel distribution.
# generates a .whl file in the dist directory
python -m build --wheel
Semantic Version Bump
Create .github/workflows/bump.yml as follows:
name: Bump
on:
release:
types: published
jobs:
bump:
runs-on: ubuntu-latest
steps:
- name: checkout
uses: actions/checkout@v4
- name: bump
id: bump
run: |
# VERSION=$(cargo tree | grep automesh | cut -d " " -f 2 | cut -d "v" -f 2)
VERSION=$(grep version pyproject.toml | cut -d '"' -f 2)
MAJOR_MINOR=$(echo $VERSION | rev | cut -d "." -f 2- | rev)
PATCH=$(echo $VERSION | rev | cut -d "." -f 1)
BUMP=$(( $PATCH + 1))
BUMPED_VERSION=$(echo $MAJOR_MINOR"."$BUMP)
BUMP_BRANCH=$(echo "bump-$VERSION-to-$BUMPED_VERSION")
echo "bump_branch=$BUMP_BRANCH" >> $GITHUB_OUTPUT
sed -i "s/version = \"$VERSION\"/version = \"$BUMPED_VERSION\"/" pyproject.toml
git config --global user.email "bump"
git config --global user.name "bump"
git add pyproject.toml
git commit -m "Bumping version from $VERSION to $BUMPED_VERSION."
git branch $BUMP_BRANCH
git checkout $BUMP_BRANCH
git push --set-upstream origin $BUMP_BRANCH
- name: pr
uses: rematocorp/open-pull-request-action@v1
with:
github-token: ${{ secrets.GITHUB_TOKEN }}
from-branch: ${{ steps.bump.outputs.bump_branch }}
to-branch: ${{ github.event.repository.default_branch }}
repository-owner: autotwin
repository: ${{ github.event.repository.name }}